
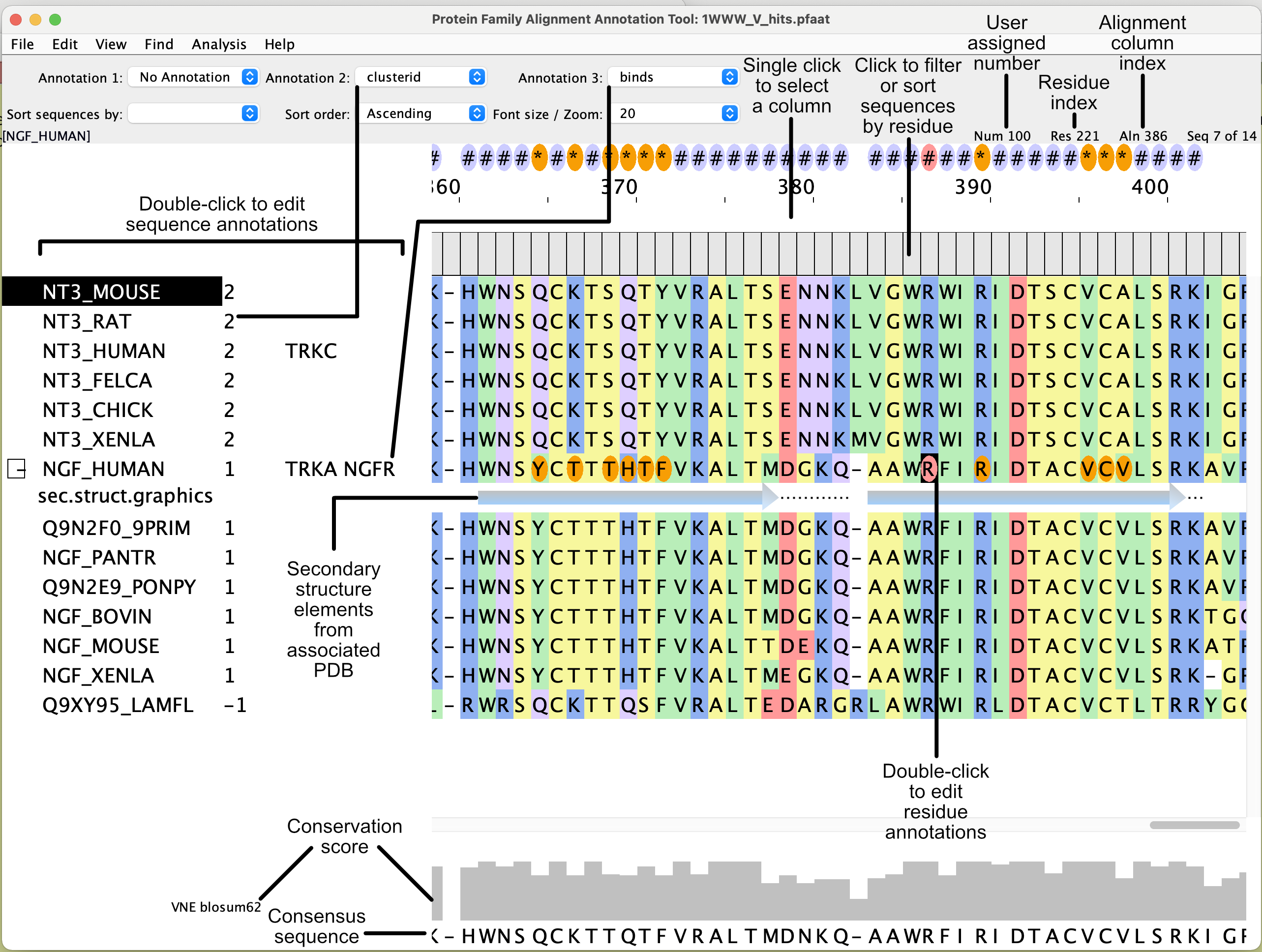
Pfaat (pronounced Pea-fat) is a Java application that allows one to edit, analyze, and annotate multiple sequence alignments. Annotations are a key feature, as they provide a framework for further sequence, structural and phylogenetic analysis.
References
PFAAT version 2.0: a tool for editing, annotating, and analyzing multiple sequence alignments. Caffrey DR, Dana PH, Mathur V, Ocano M, Hong EJ, Wang YE, Somaroo S, Caffrey BE, Potluri S, Huang ES. 2007 Oct 11;8:381 PDF
Prediction of specificity-determining residues for small-molecule kinase inhibitors. Caffrey DR, Lunney EA, Moshinsky DJ. 2008 Nov 25;9:491 PDF
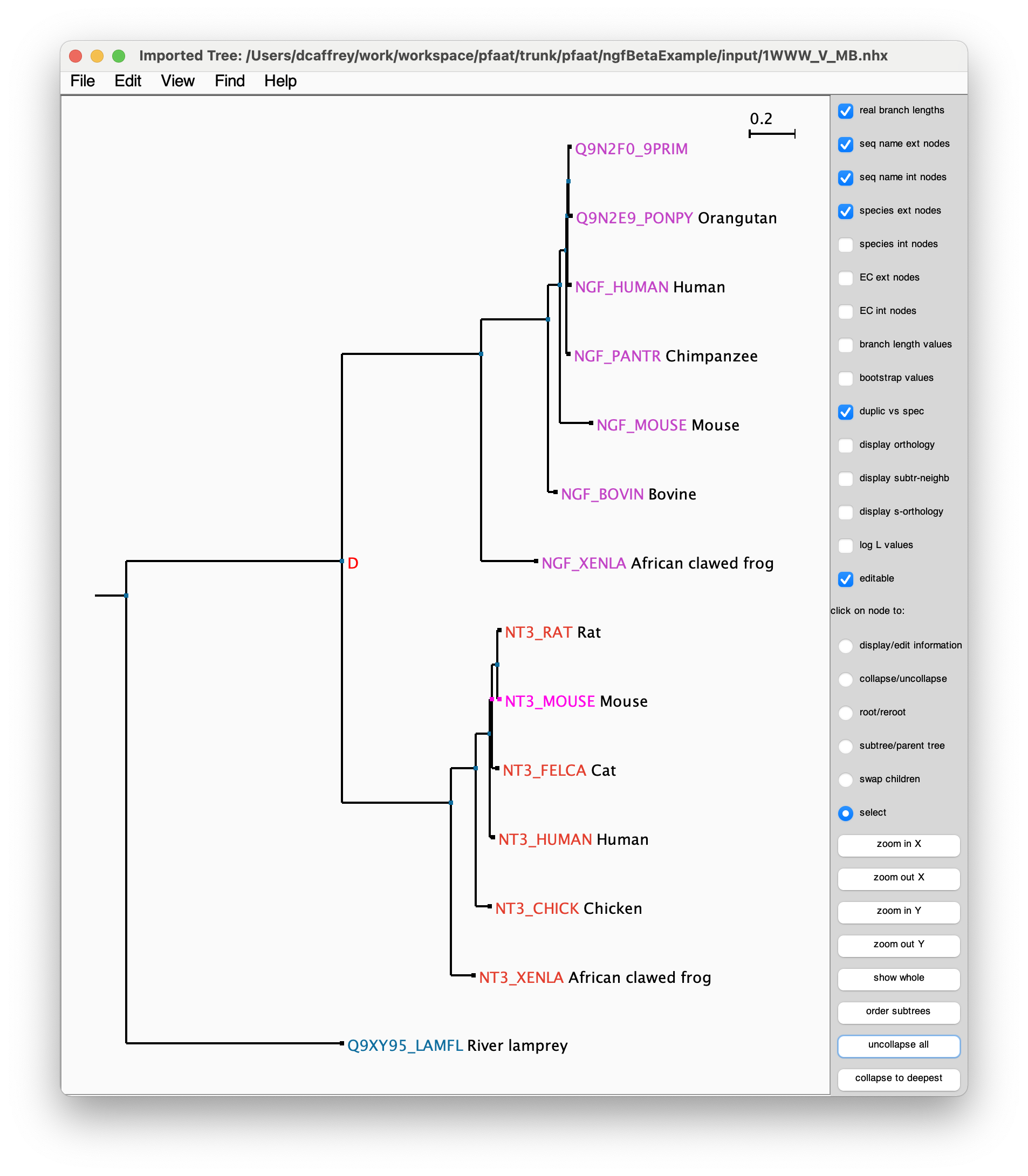
ATV: display and manipulation of annotated phylogenetic trees. Zmasek ZM, Eddy SR. 2001 Apr;17(4):383 PDF
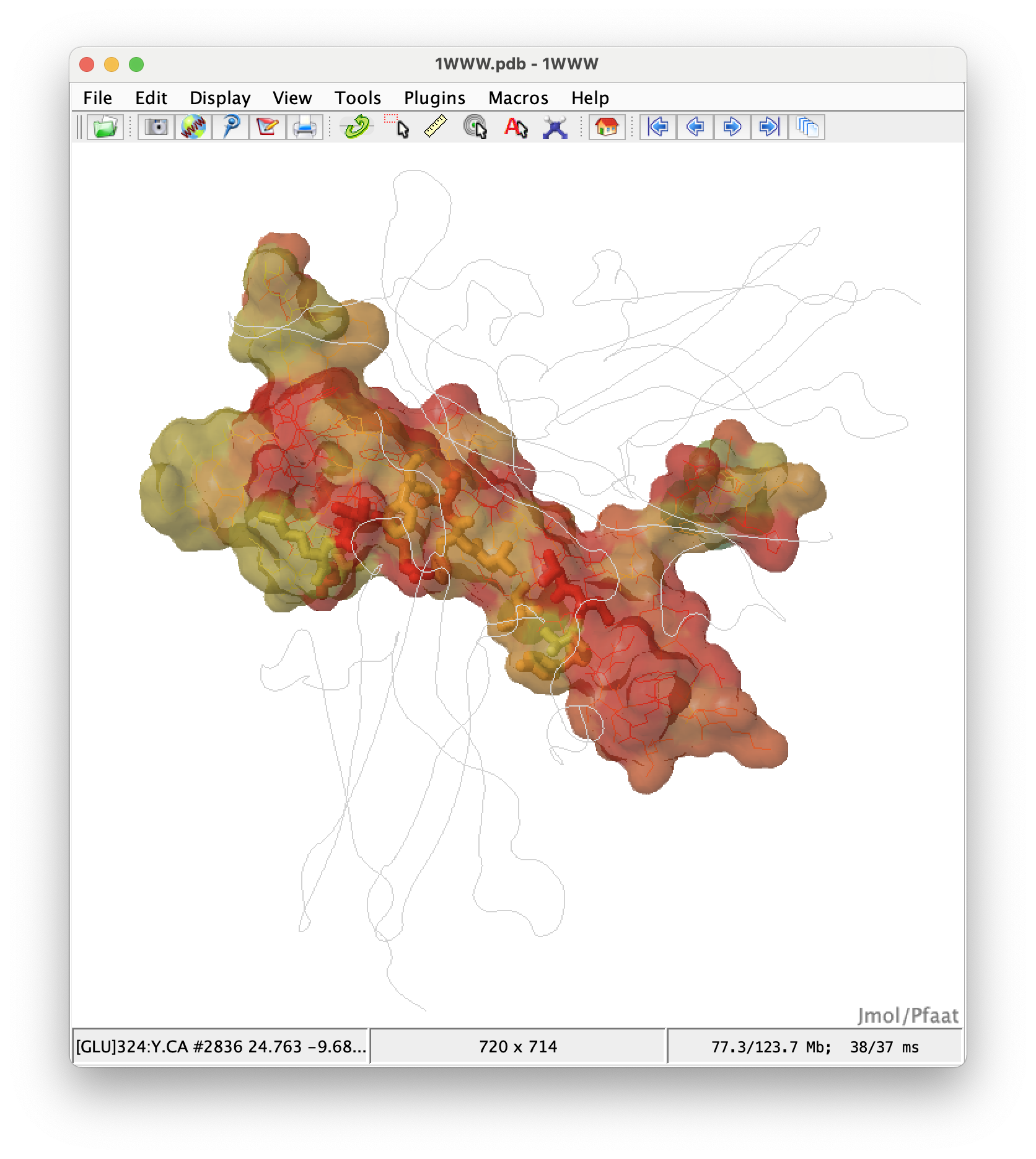
Jmol: an open-source Java viewer for chemical structures in 3D. http://www.jmol.org/